2. Monitoring Information with `impart`
impart_monitoring.RmdThis vignette demonstrates how to use impart to monitor
the amount of information contained in the accrued data so that analyses
are not under- or over-powered. This builds upon the terminology and
concepts in the vignette on study design, which can be viewed as
follows:
vignette("impart_study_design", package = "impart") # DesignMonitoring Ongoing Studies
In a fixed sample size design, the timing of the final analysis is based on the last participant’s last visit. Group sequential analyses are based when pre-specified fractions of the maximum sample size have their final outcome observed. Timing analyses in such studies only depends on counting the number of final outcomes observed during the study.
In information-monitored designs, the times at which analyses are performed and recruitment is stopped depend on when the information reaches pre-specified thresholds. The amount of information contained in the data depends on the number of observations, the completeness of the data, the analytic methods used, and the interrelationships among the observed data. Depending on the analysis methods used, an individual could contribute baseline covariates and treatment assignment information, post-randomization outcomes, and even post-randomization auxiliary variables.
Challenges in Information Monitoring
A new challenge in analyzing data in an ongoing trial is how to appropriately treat missing data. Suppose the following are observed:
- A participant is newly enrolled, has their baseline covariates measured, is randomized, is still on study, and has not yet entered any follow-up windows
- A participant is randomized, but did not attend any follow-up visits
In both of these cases, the covariates, and treatment assignment are observed, but the outcomes are missing. In the first case, outcomes are not yet observed: while they are missing, they could still be observed when the participant enters the study window. In the latter case, they are known to be missing: the study window is closed without the outcome being observed.
A convention can be used to differentiate outcomes that are not yet observed from those known to be missing:
- Completed assessments have both an observed outcome and an observed assessment time.
- Missed assessments have a missing outcome, but an observed
assessment time:
- If an assessment occurred, but no valid outcome was obtained, the time of the assessment is used.
- If an assessment was missed, the end of the study window is used, indicating the last time an outcome could have been observed per protocol.
- Not-yet-observed assessments are missing both an outcome and an assessment time.
Example Data:
Information monitoring will be illustrated using a simulated dataset
named example_1:
head(example_1)
#> .id x_1 x_2 x_3 x_4 tx y_1 y_2
#> 1 1 2.0742970 0.1971432 -0.8425884 0.2794844 0 1.591873 -4.535711
#> 2 2 0.2165473 -0.7384296 0.1315016 -1.2419134 1 NA NA
#> 3 3 0.8294726 0.4997821 1.6932555 -0.4063889 0 NA NA
#> 4 4 -1.0206893 -0.2189937 -1.7719120 0.1936013 1 1.212620 -4.533776
#> 5 5 -0.0417332 0.9282685 0.8078133 0.9317145 0 8.655326 6.970372
#> 6 6 0.7275778 1.1756811 0.0226265 -0.2556343 1 6.902055 17.381316
#> y_3 y_4 .enrollment_time .t_1 .t_2 .t_3 .t_4
#> 1 13.98543 -1.320242 2.24846 25.01538 56.62636 98.51499 133.0050
#> 2 NA NA 11.05565 55.05565 85.05565 115.05565 145.0556
#> 3 NA NA 16.96591 60.96591 90.96591 120.96591 150.9659
#> 4 11.17615 -6.629545 25.13396 59.84544 81.58577 127.78659 154.3419
#> 5 17.62329 9.126240 50.07301 75.94952 111.21967 143.00338 181.4162
#> 6 -2.42570 3.549977 50.93935 80.29181 114.96781 151.34249 177.3846The dataset is in wide format, with one row per individual. There are
four baseline covariates: x_1, x_2, …,
x_4. Treatment assignment is indicated a binary indicator
(tx), with the time from study initiation to randomization
indicated by .enrollment_time. There are four outcomes
assessed at 30, 60, 90, and 120 days post-randomization:
y_1, y_2, y_3 and
y_4. The time to each observed outcome is indicated in
columns .t_1, .t_2, .t_3 and
.t_4. For missed study visits, the time recorded is the
last day within the study window at which the individual’s outcome could
have been assessed.
Preparing Interim Datasets
A function prepare_interim_data allows the user to
create indicator variables for each outcome, with each variable
indicating whether an outcome is observed (1), is known to
be missing (0), or has not yet been observed
(NA). This allows software to appropriately handle missing
information during an ongoing study.
prepare_interim_data retains only the columns of data
relevant to the analysis at hand: covariates, study entry/enrollment
time, treatment assignment, outcomes, and the times at which outcomes
were measured.
# Obtain time of last event
last_event <-
example_1[, c(".enrollment_time", ".t_1", ".t_2", ".t_3", ".t_4")] |>
unlist() |>
max(na.rm = TRUE) |>
ceiling()
prepared_final_data <-
prepare_monitored_study_data(
data = example_1,
study_time = last_event,
id_variable = ".id",
covariates_variables = c("x_1", "x_2", "x_3", "x_4"),
enrollment_time_variable = ".enrollment_time",
treatment_variable = "tx",
outcome_variables = c("y_1", "y_2", "y_3", "y_4"),
outcome_time_variables = c(".t_1", ".t_2", ".t_3", ".t_4"),
# Observe missingness 1 week after target study visit
observe_missing_times = c(30, 60, 90, 120) + 7
)The resulting object contains the prepared dataset, the original dataset, the study time of the data lock, and a list of variables and their role in analyses.
Reverting to Information Earlier in the Study
These conventions can also be used to take a dataset from one point in time during the study, and revert to the information that was only available at an earlier point in the study. This can be useful for determining how quickly information is accruing during an ongoing study. Let , , , and respectively denote baseline covariates, treatment assignment, the outcome observed at study visit , and the times at which the study outcomes are observed. Let denote the closing of the study window for visit .
To obtain the information available at days after the randomization of the first participant:
- Retain only participants where : i.e. those randomized by study time
- Set outcome to unobserved if : i.e. outcomes not observed by time
- Set to missing if : otherwise, treat the outcome as not yet observed.
For example, the data known at 90 days can be obtained using the
data_at_time_t() function as follows:
data_90 <-
data_at_time_t(
prepared_data = prepared_final_data,
study_time = 90
)Consider 90 days after study initiation:
- Participant 1 is known to have missed their first post-randomization
assessment:
.r_1is0 - Participants 2-5 have had their first post-randomization outcome
obtained:
.r_1is1 - Participant 6 missed their first post-randomization assessment, but
this is not yet known at day 90:
.r_1isNA
show_cols <- c(".id", "x_2", "x_3", "x_4", "tx")
# Original Data
data_90$original_data[1:6, c(show_cols, ".enrollment_time", ".t_1", "y_1")]
#> .id x_2 x_3 x_4 tx .enrollment_time .t_1 y_1
#> 1 1 0.1971432 -0.8425884 0.2794844 0 2.24846 25.01538 1.591873
#> 2 2 -0.7384296 0.1315016 -1.2419134 1 11.05565 55.05565 NA
#> 3 3 0.4997821 1.6932555 -0.4063889 0 16.96591 60.96591 NA
#> 4 4 -0.2189937 -1.7719120 0.1936013 1 25.13396 59.84544 1.212620
#> 5 5 0.9282685 0.8078133 0.9317145 0 50.07301 75.94952 8.655326
#> 6 6 1.1756811 0.0226265 -0.2556343 1 50.93935 80.29181 6.902055
# Data known at study day 90
data_90$data[1:6, c(show_cols, ".e", ".t_1", "y_1", ".r_1")]
#> .id x_2 x_3 x_4 tx .e .t_1 y_1 .r_1
#> 1 1 0.1971432 -0.8425884 0.2794844 0 2.24846 25.01538 1.591873 1
#> 2 2 -0.7384296 0.1315016 -1.2419134 1 11.05565 55.05565 NA 0
#> 3 3 0.4997821 1.6932555 -0.4063889 0 16.96591 60.96591 NA 0
#> 4 4 -0.2189937 -1.7719120 0.1936013 1 25.13396 59.84544 1.212620 1
#> 5 5 0.9282685 0.8078133 0.9317145 0 50.07301 75.94952 8.655326 1
#> 6 6 1.1756811 0.0226265 -0.2556343 1 50.93935 80.29181 6.902055 1If observe_missing_times were set to 0 for
each outcome, any outcome that is NA and has a recorded
time of assessment will be treated as missing.
Plotting Observed Number of Outcomes
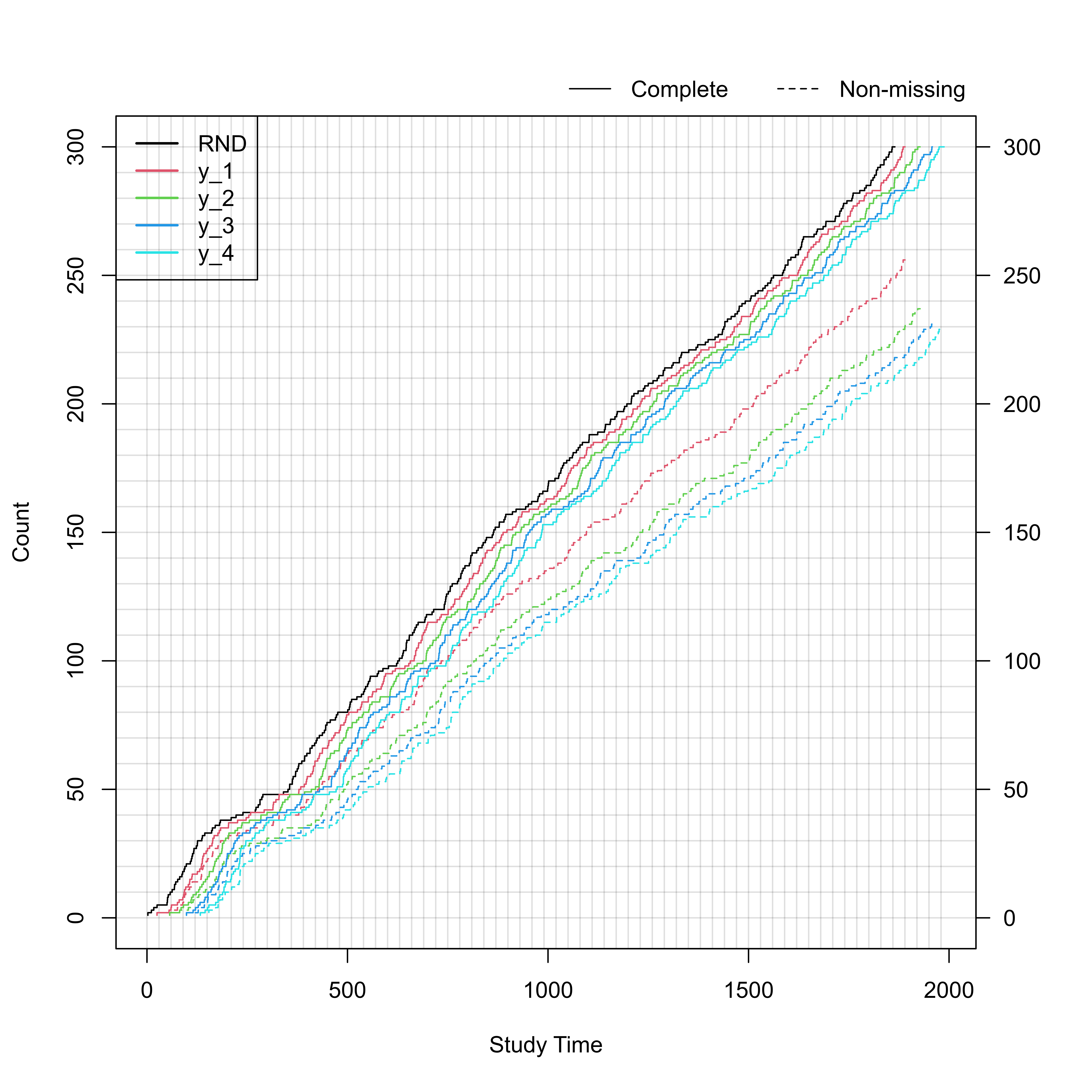
One part of monitoring involves determining how many individuals contribute a given type of information to analyses, including baseline covariates, intermediate outcomes, and final outcomes. For binary and time-to-event analyses, monitoring should include the number of observed events, not just the number of participants who have completed monitoring.
Plotting can be done for all available events:
# Plot events at the end of the study
plot_outcome_counts(
prepared_data = prepared_final_data
)
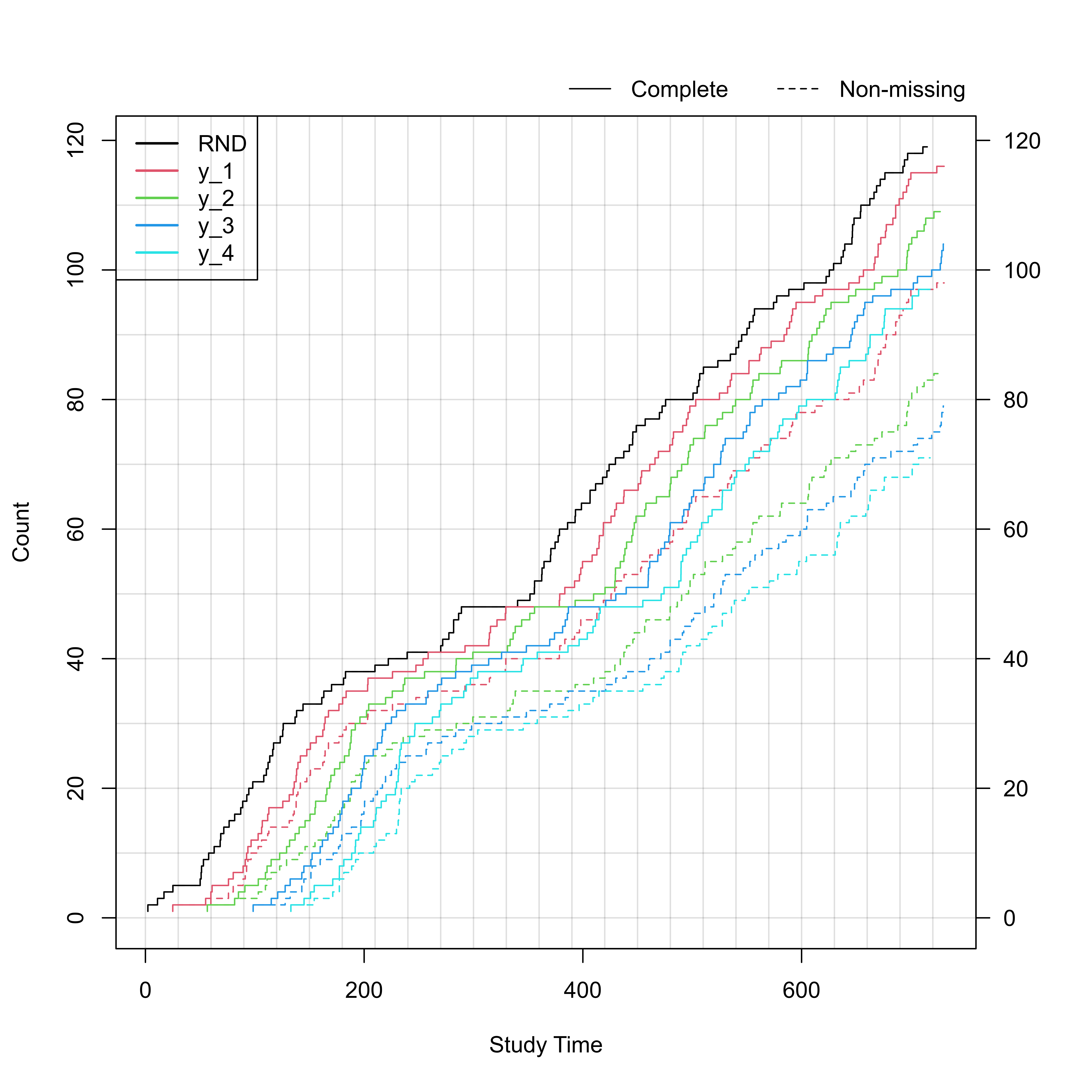
Plotting can also be done for events known at a particular point in time:
# Plot events two years into the study
plot_outcome_counts(
prepared_data = prepared_final_data,
study_time = 731 # 2 Years
)
Tabulating Event Times
The times of each event can be obtained using
count_outcomes:
example_1_counts <-
count_outcomes(
prepared_data = prepared_final_data
)
# Timing of first n randomizations
subset(
example_1_counts,
event == "randomization"
) |> head()
#> event time count_total count_complete
#> 1 randomization 2.24846 1 1
#> 2 randomization 11.05565 2 2
#> 3 randomization 16.96591 3 3
#> 4 randomization 25.13396 4 4
#> 5 randomization 50.07301 5 5
#> 6 randomization 50.93935 6 6
# Timing of first n observations of `y_4`
subset(
example_1_counts,
event == "y_4"
) |> head()
#> event time count_total count_complete
#> 1106 y_4 133.0050 1 1
#> 2106 y_4 145.0556 2 NA
#> 316 y_4 150.9659 3 NA
#> 416 y_4 154.3419 4 2
#> 816 y_4 171.3021 5 3
#> 716 y_4 177.3249 6 4
# Find when n observations of `y_4` are first available:
subset(
example_1_counts,
event == "y_4" & count_complete == 70
)
#> event time count_total count_complete
#> 944 y_4 707.0069 96 70This can also be used with data_at_time_t to reconstruct
the study data when a particular number of observations have
accrued:
# Reconstruct the data when N = 70 final outcomes were obtained
data_n_final_70 <-
data_at_time_t(
prepared_data = prepared_final_data,
study_time =
# Time when 70 final outcomes are observed:
ceiling(
subset(
example_1_counts,
event == "y_4" & count_complete == 70
)$time
)
)
data_n_70 <- data_n_final_70$dataMonitoring Information Levels
Information can be computed using the
estimate_information function: users pass the function
which conducts the analysis (estimation_function), along
with a list of parameters the function requires
(estimation_arguments). When there are multiple analyses,
the orthogonalize argument specifies whether the test
statistics and covariance should be orthogonalized to meet the
independent increments assumption. A random number generator seed
(rng_seed) can be supplied for reproducibility. By default,
only the variance and information are returned, which can be compared
against the target information level for analyses:
information_n_70 <-
estimate_information(
data = data_n_70,
monitored_design = NULL,
estimation_function = standardization,
estimation_arguments =
list(
estimand = "difference",
y0_formula = y_4 ~ x_1 + x_2 + x_3 + x_4,
y1_formula = y_4 ~ x_1 + x_2 + x_3 + x_4,
family = gaussian,
treatment_column = "tx",
outcome_indicator_column = ".r_4"
),
orthogonalize = TRUE,
rng_seed = 23456
)
information_n_70$covariance_uncorrected
#> estimates
#> estimates 4.333183
information_n_70$information
#> estimates
#> 0.2307772
information_n_70$covariance_orthogonal_uncorrected
#> estimates
#> estimates 4.333183
information_n_70$information_orthogonal
#> NULLWhen a study is designed with interim and final analyses,
monitored_design allows the user to pass the results of
previously conducted analyses to estimate_information.
Using the design specified in the impart_study_design
vignette:
# Universal Study Design Parameters
minimum_difference <- 5 # Effect Size: Difference in Means of 5 or greater
alpha <- 0.05 # Type I Error Rate
power <- 0.9 # Statistical Power
test_sides <- 2 # Direction of Alternatives
# Determine information required to achieve desired power at fixed error rate
information_single_stage <-
impart::required_information_single_stage(
delta = minimum_difference,
alpha = alpha,
power = power
)
# Group Sequential Design Parameters
information_rates <-
c(0.50, 0.75, 1.00) # Analyses at 50%, 75%, and 100% of the Total Information
type_of_design <- "asOF" # O'Brien-Fleming Alpha Spending
type_beta_spending <- "bsOF" # O'Brien-Fleming Beta Spending
# Set up group sequential testing procedure
trial_design <-
rpact::getDesignGroupSequential(
alpha = alpha,
beta = 1 - power,
sided = 2,
informationRates = information_rates,
typeOfDesign = type_of_design,
typeBetaSpending = type_beta_spending,
bindingFutility = FALSE
)
# Inflate information level to account for multiple testing
information_adaptive <-
impart::required_information_sequential(
information_single_stage = information_single_stage,
trial_design = trial_design
)
# Initialize the monitored design
monitored_design <-
initialize_monitored_design(
trial_design = trial_design,
null_value = 0,
maximum_sample_size = 280,
information_target = information_adaptive,
orthogonalize = TRUE,
rng_seed_analysis = 54321
)The current information fraction can be computed:
information_n_70$information/information_adaptive
#> estimates
#> 0.5062808Tools for Monitoring Information
While estimate_information can provide an estimate of
the information at a particular point in the study, understanding the
rate at which information is accruing over time can be useful in
projecting when pre-specified information levels may be met:
data_n_70_trajectory <-
information_trajectory(
prepared_data = data_n_final_70,
monitored_design = monitored_design,
estimation_function = standardization,
estimation_arguments =
list(
estimand = "difference",
y0_formula = y_4 ~ x_1 + x_2 + x_3 + x_4,
y1_formula = y_4 ~ x_1 + x_2 + x_3 + x_4,
family = gaussian,
treatment_column = "tx",
outcome_indicator_column = ".r_4"
),
correction_function = standardization_correction,
orthogonalize = TRUE,
n_min = 40,
n_increment = 2,
rng_seed = 23456,
control = monitored_analysis_control()
)
#> Error in information$information[i] <- if (orthogonalize) {: replacement has length zero
data_n_70_trajectory
#> Error in eval(expr, envir, enclos): object 'data_n_70_trajectory' not foundThis trajectory can be smoothed using regression, ideally using a
method resistant to outliers, such as deming::theilsen.
Inverse regression can be used to obtain an estimated number of outcomes
necessary to achieve a given level of information:
plot(
information_fraction ~ y_4,
data = data_n_70_trajectory,
ylim = c(0, 1)
)
#> Error in eval(m$data, eframe): object 'data_n_70_trajectory' not found
abline(
lm(
formula = information_fraction ~ y_4,
data = data_n_70_trajectory
),
lty = 1
)
#> Error in eval(mf, parent.frame()): object 'data_n_70_trajectory' not found
# Requires `deming` package
abline(
deming::theilsen(
formula = information_fraction ~ y_4,
data = data_n_70_trajectory
),
lty = 3
)
#> Error in eval(temp, parent.frame()): object 'data_n_70_trajectory' not found
abline(
h = monitored_design$original_design$information_fractions,
lty = 2
)
#> Error in int_abline(a = a, b = b, h = h, v = v, untf = untf, ...): plot.new has not been called yet